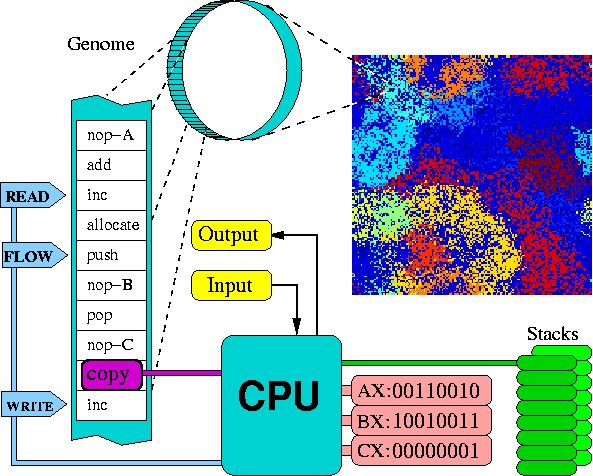
This document describes the structure of the classic virtual CPU and an example organism running on it.
The virtual CPU, which is the default "body" or "hardware" of the organisms, contains the following set of components, (as further illustrated in the figure below).

The instruction set in Avida is loaded on startup from a configuration file specified in the avida.cfg file. This allows selection of different instruction sets without recompiling the source code, as well as allowing different sized instruction sets to be specified. It is not possible to alter the behavior of individual instructions or add new instructions without recompiling Avida; such activities have to be done directly in the source code.
The available instructions are listed in the inst_set.* files with
a 1 or a 0 next to an instruction to indicate if it
should or should not be included. Changing the instruction set to be used
simply involves adjusting these flags.
The instructions were created with three things in mind:
One major concept that differentiates this virtual assembly language from its
real-world counterparts is in the additional uses of nop instructions
(no-operation commands).
These have no direct effect on the virtual CPU when executed, but often
modify the effect of any instruction that precedes them. In a sense, you
can think of them as purely regulatory genes. The default instruction set
has three such nop instructions: nop-A, nop-B,
and nop-C.
The remaining instructions can be seperated into three classes. The first
class is those few instructions that are unaffected by nops. Most of these
are the "biological" instructions involved directly in the replication
process. The second class of instructions is those for which a nop
changes the head or register affected by the previous command. For example,
an inc command followed by the instruction nop-A would
cause the contents of the AX register to be incremented, while an
inc command followed by a nop-B would increment BX.
The notation we use in instruction definitions to describe that a default
component (that is, a register or head) can be replaced due to a nop command
is by surrounding the component name with ?'s. The component listed is the
default one to be used, but if a nop follows the command, the
component it represents in this context will replace this default.
If the component between the question marks is a register than a
subsequent nop-A represents the AX register,
nop-B is BX, and nop-C is CX.
If the component listed is a head (including the instruction pointer) then a
nop-A represents the Instruction Pointer, nop-B represents
the Read-Head, and nop-C is the Write-Head. Currently
the Flow-Head has no nop associated with it.
The third class of instructions are those that use a series of nop
instructions as a template (label) for a command that needs to reference
another position in the code, such as
h-search. If nop-A follows a search command, it scans for
the first complementary template (nop-B) and moves the Flow-Head
there. Templates may be composed of more than a single nop
instruction. A series of nops is typically abbreviated to the associated
letter and separated by colons. This the sequence "nop-A nop-A nop-C"
would be displayed as "A:A:C".
The label system used in Avida allows for an arbitrary number of nops. By
default, we have three: nop-A's complement is nop-B,
nop-B's is nop-C, and nop-C's is nop-A.
Likewise, some instructions talk about the complement of a register or head
-- the same pattern is used in those cases. So if an instruction tests if
?BX? is equal to its complement, it will test if
BX == CX by default, but if it is followed by a
nop-C it will test if CX == AX.
An abbreviated description of the 26 default instructions is below.
| (a-c) | nop-A, nop-B, and nop-C
| No-operation instructions; these modify other instructions. |
|---|---|---|
| (d) | if-n-equ
| Execute next instruction only-if ?BX? does not equal its complement |
| (e) | if-less
| Execute next instruction only if ?BX? is less than its complement |
| (f) | if-label
| Execute the next instruction only if the given template complement was just copied |
| (g) | mov-head
| Move the ?IP? to the same position as the Flow-Head |
| (h) | jmp-head
| Move the ?IP? by a fixed amount found in CX |
| (i) | get-head
| Write the position of the ?IP? into CX |
| (j) | set-flow
| Move the Flow-Head to the memory position specified by ?CX? |
| (k) | shift-r
| Shift all the bits in ?BX? one to the right |
| (l) | shift-l
| Shift all the bits in ?BX? one to the left |
| (m) | inc
| Increment ?BX? |
| (n) | dec
| Decrement ?BX? |
| (o) | push
| Copy the value of ?BX? onto the top of the current stack |
| (p) | pop
| Remove a number from the current stack and place it in ?BX? |
| (q) | swap-stk
| Toggle the active stack |
| (r) | swap
| Swap the contents of ?BX? with its complement. |
| (s) | add
| Calculate the sum of BX and CX; put the result in ?BX? |
| (t) | sub
| Calculate the BX minus CX; put the result in ?BX? |
| (u) | nand
| Perform a bitwise NAND on BX and CX; put the result in ?BX? |
| (v) | h-copy
| Copy an instruction from the Read-Head to the Write-Head and advance both. |
| (w) | h-alloc
| Allocate memory for an offspring |
| (x) | h-divide
| Divide off an offspring located between the Read-Head and Write-Head. |
| (y) | IO
| Output the value ?BX? and replace it with a new input |
| (z) | h-search
| Find a complement template and place the Flow-Head after it. |
| # --- Setup --- | |
h-alloc | # Allocate extra space at the end of the genome to copy the offspring into. |
| h-search | # Locate an
A:B template (at the end of the organism) and
place the Flow-Head after it |
| nop-C | # |
| nop-A | # |
| mov-head | # Place the
Write-Head at the Flow-Head (which is at beginning of
offspring-to-be). |
| nop-C | # [ Extra
nop-C commands can be placed here w/o harming the
organism! ] |
| # --- Copy Loop --- | |
| h-search | # No template,
so place the Flow-Head on the next line code |
h-copy | # Copy a single
instruction from the read head to the write head (and advance both
heads!) |
if-label | # Execute the
line following this template only if we have just copied an
A:B template. |
nop-C | # |
nop-A | # |
h-divide | # ...Divide off offspring! (note if-statement above!) |
mov-head | # Otherwise,
move the IP back to the Flow-Head at the beginning of the copy
loop. |
nop-A | # End label. |
nop-B | # End label. |
This program begins by allocating extra space for its offspring. The exact amount of space does not need to be specified -- it will allocate as much as it is allowed to. The organism will then do a search for the end of its genome (where this new space was just placed) so that it will know where to start copying. First the Flow-Head is placed there, and then the Write-Head is moved to the same point.
It is after this initial setup and before the actual copying process
commences that extra nop instructions can be included. The only
caveat is that you need to make sure that you don't duplicate any templates
that the program will be searching for, or else it will no longer function
properly. The easiest thing to do is insert a long sequence of nop-C
instructions.
Next we have the beginning of the "copy loop". This segement of code starts
off with an h-search command with no template following it. In
such as case, the Flow-Head is placed on the line immediately following the
search. This head will be used to designate the place that the IP keeps
returning to with each cycle of the loop.
The h-copy command will copy a single instruction from the Read-Head
(still at the very start of the genome, where it begins) to the Write-Head
(which we placed at the beginning of the offspring). With any copy command
there is a user-specified chance of a copy mutation. If one occurs, the
Write-Head will place a random instruction rather than the one that it
gathered from the Read-Head. After the copy occurs (for better or worse),
both the Read-Head and the Write-Head are advanced to the next instruction
in the genome. It is for this reason that a common mutation we see happening
will place a long string of h-copy instruction one after another.
The next command, if-label (followed by a nop-C and a
nop-A) tests to see if the complement of C:A is the most
thing copied. That is, if the two most
recent instructions copied were a nop-A followed by a nop-B
as is found at the end of the organism. If so, we are done! Execute the
next instruction which is h-divide (when this occurs, the read and
write heads will surround the portion of memory to be split off as the
offspring's genome). If not, then we need to keep
going. Skip the next instruction and move on to the mov-head which
will move the head specified by the nop that follows (in this case
nop-A which is the Instruction Pointer) to the Flow-Head at the
beginning of the copy loop.
This process will continue until all of the lines of code have been copied, and an offspring is born.
Here is a short example program to demonstrate one way for an organism to perform the "OR" logic operation. This time I'm only going to show the contents of the registers after each command because the functionality of the individual instructions should be clear, and the logic itself won't be helped much by a line-by-line explanation in English.
| Line # | Instruction | AX | BX | CX | Stack | Output |
|---|---|---|---|---|---|---|
| 1 | IO | ? | X | ? | ? | ? |
| 2 | push | ? | X | ? | X, ? | |
| 3 | pop | ? | X | X | ? | |
| 4 | nop-C | |||||
| 5 | nand | ~X | X | X | ? | |
| 6 | nop-A | |||||
| 7 | IO | ~X | Y | X | ? | X |
| 8 | push | ~X | Y | X | Y, ? | |
| 9 | pop | ~X | Y | Y | ? | |
| 10 | nop-C | |||||
| 11 | nand | ~X | ~Y | Y | ? | |
| 12 | swap | Y | ~Y | ~X | ? | |
| 13 | nop-C | |||||
| 14 | nand | Y | X or Y | ~X | ? | |
| 15 | IO | Y | Z | ~X | ? | X or Y |